|
 |
|||||||||||
|
|
|||||||||||
|
Evolution of IC: Evolution of Hormone-Receptor ComplexityBy Ian MusgravePosted April 14, 2006 Update: Ive added some substantial comments 95979 and 96005 that might be lost in the comment roll, but add some important perspective to Behe's arguments. Michael Behe is known as the author of the concept of Irreducible Complexity (IC, but see [note 1]). However, he has given several different, not entirely consistent, definitions of IC. Everyone is familiar with the "multiple parts" definition, fewer will be familiar with the "neutral mutational steps" definition (1) and fewer still with the idea that amino acids interactions themselves are IC (2, see my critique of this). Indeed, Behes recent paper with David Snoke (3) relied on a combination of the last two definitions (see our critique), and Behe also used the latter definition in the Dover trial (3). A paper just out in the journal Science has effectively refuted the claims of the Behe and Snoke paper (4) Behe has claimed that he will accept that a system is not IC if someone can give a mutation-by-mutation account, with selective coefficients for each mutation, for that system. Now in general, this is just not practical, because it is very difficult to reconstruct these systems even for bacterial antibiotic resistance, where we can keep a close eye on the mutations, and can readily measure the effect of the mutations. In fact, for resistance to cefotaxamine, 5 mutations are required, and there are 18 high probability pathways to this resistance (5), so we have little idea of the historical path the system took. Reconstructing the actual mutation-by-mutation path of any given feature, especially in organisms that are not as easy to grow or test as bacteria (and given that many intermediate organisms may be extinct), is a daunting task. By requesting a ludicrous amount of detail, Behe has tried to insulate himself from the evidence for evolution. In a magnificent paper, Bridgham, Carroll and Thornton (BCT, 4) have tackled the daunting task of reconstructing evolutionary pathways head on, reconstructing ancient proteins in the process. The article looks at one of Behes key examples, protein-binding sites for small molecules. The ability of proteins to bind small molecules is critical for organisms to function, because enzymes bind small molecules as part of metabolism, receptors proteins bind small hormone molecules to initiate cell signaling and many small molecules bind to a variety of proteins to modify their functions. For example, in the Behe and Snoke paper (3), they examine the ability of the oxygen carrying protein haemoglobin to bind the organic phosphate molecule 2,3-diphosphoglycerate (DPG, which modifies the oxygen binding capacity of haemoglobin). The BCT paper looks at the binding site of the receptors for some steroid hormone receptors, those for mineralocorticoids (MR) and those for glucacorticoids (GR). Receptors are proteins whose amino acid chains fold in such a way as to produce a pocket where particular small molecules (hereafter called ligands) bind. These pockets can be incredibly selective, and the term "lock and key" is often used. The small molecule fits into the protein receptor like a key into a lock (this is an oversimplification, as the molecules are flexible, and are "floppy" keys and locks, and the electrostatic charge and lipid solubility of the molecule comes into play, but it helps visualization).
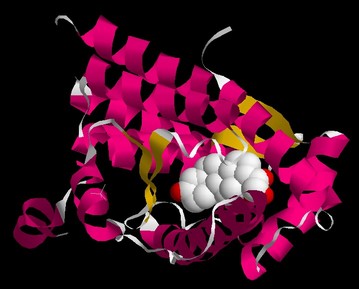
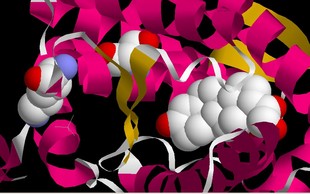
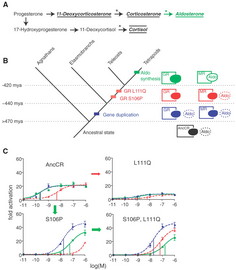
Now, modern tetrapods (amphibians, reptiles, birds and mammals) have separate receptors for the steroid hormones cortisol (GR, which modulates metabolism, inflammation and immunity) and aldosterone (MR, modulates salt balance amongst other things). Hagfish and Lampreys ("primitive" jawless fish with cartilaginous skeletons) have only one receptor, which is activated by both aldosterone and cortisol. Sharks and such have two receptors, both of which are activated by both aldosterone and cortisol. Finally, bony fish and tetrapods have two receptors, one which is activated by aldosterone, and one which is activated by cortisol. What were the molecular steps which brought this about? The aldosterone receptor (as helix and strings) showing aldosterone (the white collection of balls) binding in the pocket. On the right is a close-up, with the amino acids that are mutated to make the GR receptor shown as balls as well. BCT approach this in a very elegant way. Using phylogenetic analysis of the existing sequences of GR and MR receptors they were able to reconstruct the sequence of the receptor ancestral to the GR and MR. They expressed this reconstructed gene in cells, and tested the sensitivity to aldosterone and cortisol. The ancestral receptor responded to both aldosterone and cortisol. By a combination of phylogenetic analysis and mutagenesis they isolated two mutations that converted the ancestral receptor into a GR (serine to proline at position 106 in the chain, and lysine to glutamine at position 111). By studying their properties, and comparing them to MR and GR receptors from hagfish, sharks, bony fish and tetrapods, they determined that the seriene to proline mutation came first, followed by the lysine to glutamate.
Deriving the ancestral sequence, determining the mutations, their temporal sequence and their relative selectability is all in all a virtuoso performance. And it directly addresses the issue Behe raised with the evolution of the DPG binding site (3, see our critique). Behe and the Discovery Insitute have reacted quickly and negatively to this paper. But in doing so they display a curious amnesia. Behe says:
Yet this "system" is precisely the thing that Behe uses in his exemplar for the Behe and Snoke paper, the binding of DPG to haemoglobin. And Behe has said in testimony to the Dover trial (3) that the Behe and Snoke paper on evolution of binding sites is about irreducible complexity. So if the evolution of the DPG binding site (where you only need two mutations to make a functioning DPG binding site) is an example of IC, then the evolution of the aldosterone binding site is also (note 2). As the BCT paper specifically cites the Behe and Snoke paper, you would expect they would look at the ideas contained in the paper, not "Darwins Black Box". Behe has had a long history of citing examples of molecular IC. He has even called disulfide bond "irreducibly complex" (2). So his disavowal of an example that directly addresses the Behe and Snoke paper (3) is particularly disingenuous. Stephen Meyer also argues on this line.
Yet, and I emphasise this again, Behe himself has argued in sworn testimony to the Dover trial that they do constitute IC systems (3). Don't the DI fellows follow each others work? Once again we see that the Intelligent design promoters are willing to move the goal posts to avoid refutation of their ideas. Molecular IC is Behes own invention, and he cant ignore his idea when it is disproved. Notes and references. [Note 1] Actually, evolutionary biologist H.J. Muller came up with irreducible complexity in 1918, as a prediction of Darwinian evolution. See H. J. Muller, "Reversibility in Evolution Considered from the Standpoint of Genetics," Biological Reviews 14 (1939): 261-80. In 1986 another evolutionary biologist, Cairns-Smith, also described IC systems produced by evolutionary processes. [Note 2] Technically, the GR receptor is an example of subfunctionalisation, where a generalist receptor becomes more selective. This is quite important in evolution. Behes exemplar, binding DPG in haemoglobin, is a result of a reduction of specificty, the ATP binding site now binds DPG. When Meyer complains that the BCT paper does not generate a new protein fold family, well, neither does Behes DPG example, which Behe claims demonstrates IC. (ironically, there is also a recent paper showing in more detail how protein fold familes evolve. Zeldovich KB, Berezovsky IN, Shakhnovich EI. Physical origins of protein superfamilies. J Mol Biol. 2006 Apr 7;357(4):1335-43.) (1) "An irreducibly complex evolutionary pathway is one that contains one or more unselected steps (that is, one or more necessary-but-unselected mutations). The degree of irreducible complexity is the number of unselected steps in the pathway." http://www.arn.org/docs/behe/mb_indefenseofblood... (2)
http://www.discovery.org/scripts/viewDB/index.ph... (3) Behe MJ & Snoke DW (2004) Simulating evolution by gene duplication of protein features that require multiple amino acid residues. Protein Science.
http://www.talkorigins.org/faqs/dover/day11pm2.h...
http://www.talkorigins.org/faqs/dover/day10pm2.h... (4) Bridgham JT, Carroll SM, Thornton JW. Evolution of hormone-receptor complexity by molecular exploitation. Science. 2006 Apr 7;312(5770):97-101. [PubMed ] [Abstract] [Full Text (subscribers only)] See also commentary Adami C. Evolution. Reducible complexity. Science. 2006 Apr 7;312(5770):61-3. [summary] [Full Text (subscribers)] (5) Weinreich DM, Delaney NF, Depristo MA, Hartl DL. Darwinian evolution can follow only very few mutational paths to fitter proteins. Science. 2006 Apr 7;312(5770):111-4. [PubMed] [Abstract] [Full text (subscribers only)] [I corrected a few typos -- Nick] Originally posted at The Panda's Thumb |